Comparative genomics of AIDS-resistant non-human primates
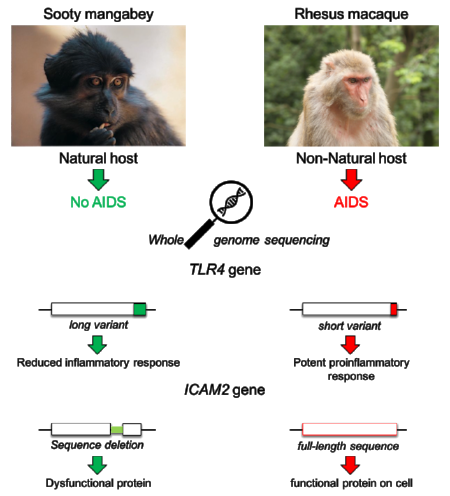
SIV infection of natural hosts, such as sooty mangabeys (Cercocebus atys, SM), is typically non-pathogenic despite high viremia. This is in stark contrast to HIV infection in humans and experimental SIV infection in rhesus macaques (Macaca mulatta, RM) that progress to AIDS unless treated with antiretroviral therapy. Over the past 15 years, the main virological and immunological features of natural SIV infection in SMs have been described in studies that compared and contrasted this infection with the pathogenic infections of HIV and SIV in humans and RMs. These studies have demonstrated that non-pathogenic infection is dependent on maintenance of low levels of immune activation during the chronic phase of the infection, downregulation of the interferon (IFN) system after infection, and the ability to achieve compartmentalization of virus replication that preserves central- and stem-cell memory CD4+ T cells as well as follicular T helper cells. Despite this progress, however, the ultimate molecular mechanisms responsible for the lack of pathogenicity in natural SIV hosts remain poorly defined. Elucidation of AIDS resistance factors in natural host species would provide remarkable insight into the molecular sequelae that drive pathogenesis and provide tractable drug targets for treating HIV-associated immune dysfunction.
In previous studies, we have used high throughput transcriptomics to characterize SIV infection in SMs and other natural hosts. These studies have identified the IFN system as a critical determinant of pathogenesis, and have initiated a series of studies designed to test the clinical utility of targeting the IFN system. With the advent of next-generation sequencing technology, the ability to study non-human primates at a genome-wide level has undergone a remarkable explosion in recent years, and draft assemblies for several species are now available. We sequenced the genome of the Sooty Mangabey and developed a novel pipeline to identify novel factors regulating primate lentiviral pathogenesis. This enabled us to identify a C-terminal frameshift in the TLR4 gene of SMs that results in an elongated C-terminus. This frameshift in TLR4 was also found in several other African natural SIV hosts such African green monkeys (AGM), colobus monkeys and drills. Subsequent analyses determined this mutation to render sooty mangabeys less responsive to bacterial LPS, thought to be a major driver of AIDS pathogenesis. Future studies will use 3rd Generation sequencing technology to sequence the genomes of additional African natural host NHPs.
Relevant Publications
Sooty mangabey genome sequence provides insight into AIDS resistance in a natural SIV host (Nature)
Natural SIV hosts: showing AIDS the door (Science)
Nonpathogenic SIV Infection of Sooty Mangabeys (Encyclopedia of AIDS)
Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque. (J Clin Invest)
Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. (J Clin Invest)
Primate genomes for biomedicine. (Nat Biotechnol)
Systems biology of natural simian immunodeficiency virus infections. (Curr Opin HIV AIDS)